Everyone experiences itching. The sensation of Itching can result from something as innocuous as an insect bite to a more serious chronic debilitating form that can be difficult to treat.
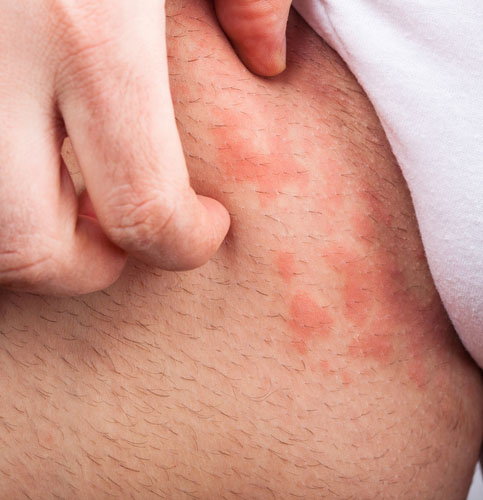
Recent advances in understanding the itching process may help in developing more effective treatments to control itch. It has been known for some time that an irritation to the skin caused by a mosquito bite or poison ivy, for example, results in an immune response by the body releasing histamine. Histamine binds to and activates the TRPV1 receptor found on the endings of sensory nerves in the skin, resulting in the transmission of a nerve signal to the brain and the sensation of itch. This understanding is the basis for the use of antihistamines for treating itch. Realizing that not all itch conditions are helped by histamines, researchers set out to learn more about the causes of itch.
A few years ago, another itch receptor (TRPA1) was discovered that is activated by substances other than histamine. Is there a link between itch and pain? The findings indicate that neurons containing only the TRPV1 receptor process pain sensation. On the other hand, neurons containing either the TRPV1 receptor or the TRPA1 receptor can transmit itch signals. The results also suggest that pain circuits can inhibit itch circuits, so only one signal is sent at a time—explaining why pain and itch rarely happen simultaneously.
Should you scratch your itch? Apparently scratching an itch has an evolutionary basis–to remove threatening bugs and plants. Scratching stimulates nerve endings in the spinal cord to release natural painkilling molecules. Scratching may bring temporary relief, but continued scratching may injure the skin leading to a greater sensation of itching.
References
1. National Institute of Arthritis and Musculoskeletal and Skin Diseases. “Investigating the Causes of Chronic Itch: New Advances Could Bring Relief.” Spotlight on Research 2014.
http://www.niams.nih.gov/News_and_Events/Spotlight_on_Research/2014/chronic_itch.asp
2. Piergrossi, Joseph. “Untangling the Source of Ouch and Itch.” National Institutes of Health, June 12, 2013. NIH article on Itch
https://publications.nigms.nih.gov/insidelifescience/untangling-ouch-itch.html
3.Sutherland, Stephani. The Maddening Sensation of Itch. Scientific American, May 2016, p. 39-43.
A New Direction- This posting concludes my blog on “keeping abreast on developments in medical research” in order to devote more time to writing a new book on asthma. The blog page on my website will be replaced by a Resources page providing brief discussions of research sources I use as background information in preparing writing assignments. These resources could be valuable to you as well in researching medical topics!